By Laurence Clarke
Insects are the most diverse group of animals on the planet, playing important roles as disease carriers, pollinators, and agricultural pests. So, when we want to know what insects are spreading disease, why certain plants are growing (or why they aren’t), or even for  conservation, it’s important that we can identify insect species accurately. I’ve recently developed new tools to identify insects using their DNA.
conservation, it’s important that we can identify insect species accurately. I’ve recently developed new tools to identify insects using their DNA.
Scientists usually identify insects visually, using morphological features like the size, shape and colour of body parts. This requires trained experts and is costly and labour intensive. On top of this, some specimens, like eggs, larvae, or that bug that turned into a smear on your windscreen, may simply be impossible to identify by morphology. An alternative is to use DNA barcodes to identify insect species, an approach that has taken off in the last 10 years.

The Monarch and Viceroy butterflies (respectively) demonstrating why looks alone can make identification difficult. Credit: R.Butler
A DNA barcode is a piece of DNA present in the genomes of all the species of interest (in this case, insects) that has enough differences in the DNA sequence such that each species is unique, allowing them to be identified. The DNA barcode for most animals, including insects, is a 650 base pair (bp) region of the mitochondrial cytochrome c oxidase I (COI) gene. This gene plays a key role in aerobic metabolism (that’s how we get energy from food).
Usually, an environmental sample like soil will contain DNA from lots of different species. Using DNA barcodes to identify multiple species from an assortment of DNA is called ‘metabarcoding’. Unfortunately, finding the long 650 bp of COI is difficult in environmental DNA samples as DNA is quickly broken down to less than 200 bp once it’s outside the cell. Even if we could find it, most machines can’t sequence 650 bp-long DNA fragments yet (although… see here for a breakdown of the current technologies). Hence, we need shorter barcodes for metabarcoding studies, ones that can be sequenced and are found in environmental samples.
Many researchers have designed markers that target smaller parts of the COI gene to try to get around this problem, but how well do the different metabarcodes work?
These days, so many DNA sequences have been stored in databases that we can test whether metabarcodes work on particular species without setting foot in the lab. For example, there are almost 2 million insect DNA barcodes in the Barcode of Life Database, aka BOLD, representing more than 60 000 species. Testing all the markers for that many insects in the lab would take a long (long, long) time. Luckily for me, a computer can run these same tests in a matter of minutes.
The computer runs established that most of the markers for detecting these shorter COI barcodes work well on flies, moths and butterflies, but fail miserably for a lot of other insects. In other words, using COI barcodes for metabarcoding would bias your results in favour of detecting some species and not others. You wouldn’t be able to tell whether certain species are present or not, so you won’t necessarily get a true picture of what’s there.
To improve detection, I designed markers targeting a completely different part of the mitochondrial DNA, namely the 16S ribosomal RNA gene. Ribosomal RNA forms stem and loop structures, where the sequence in the ‘stems’ needs to pair perfectly with another part of the RNA molecule, while that of the loops do not. This means that, unlike genes coding for proteins (such as COI), stretches of the ribosomal gene that form stems are well conserved across a wide range of insects, making them ideal targets for barcoding.
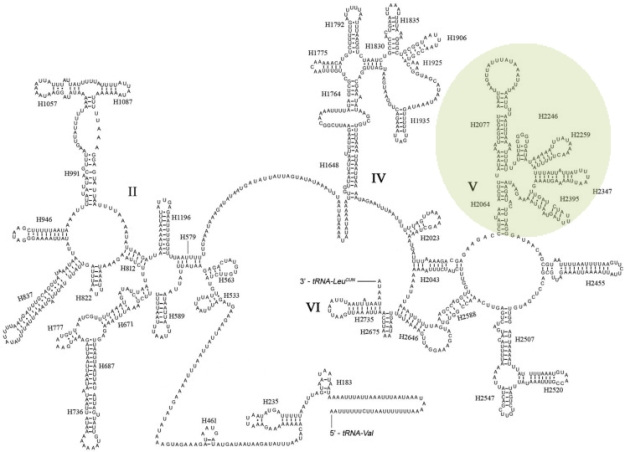
The 16S ribosomal RNA forms this complex pattern due to the stem and loop structures. My primers target the section highlighted in green. Adapted from: Wong et al.
Rerunning the computer simulations using the new 16S markers showed they worked for 95 – 97 % of the insects tested, typically amplifying more than 90 % of any given order of insects. As the coup de grace, I made an artificial blend of insect DNA in the lab and ran a metabarcoding analysis on it to see how well the new 16S markers worked. Sure enough, the 16S markers detected as many of the insect species in the blend, if not more, than the best COI markers.
So next time you want to know what that bug on your windscreen was*, give 16S a try…
For an extended read, my research was published in Molecular Ecology Resources, which you can find here.
*Pond et al. already used DNA to identify insects from ‘windshield splatter’, but they didn’t use COI or 16S. I’m still jealous that they got to it first.
